What leads to more spike protein in the body: the vaccine or virus? Part 1
A reply to an article by Uri Manor and Jeremy Howard
Background
The SARS-CoV-2 virus has a protein on its surface called the spike protein. The COVID vaccines available in the U.S. work by getting the body to produce this protein (with some modifications).
In a previous article, I went over multiple pieces of evidence showing that the spike protein alone, either from the virus or vaccine, is harmful.
A few people responded to that article by bringing up Uri Manor, an Assistant Research Professor at the Salk Institute, and senior co-author of a paper by Lei et al., which was one of the studies showing that spike protein was harmful. This paper was also one of the studies I linked to and briefly discussed in my previous article about the spike protein.
After the Lei et al. paper came out, some said that it was being used to spawn “anti-vax discussions”:
In Manor’s reply, he said that “the (relatively) small amount of spike protein produced by the mRNA vaccine would not be nearly enough to do any damage.”
Later on, Manor cited a study by Ogata et al., which actually measured the amount of free spike protein in the plasma of Moderna vaccine recipients:

Manor said that the Ogata study showed that the amount of free spike was ~100,000 times less than the paper he was a co-author on (the one which showed harmful effects of the spike protein).
After that, he and Jeremy Howard produced this article:
SARS-CoV-2 spike protein impairment of endothelial function does not impact vaccine safety
In their article, Manor and Howard don’t seem to deny the possibility that the spike protein from vaccines is harmful, but argue that the amount of spike protein produced from them is physiologically negligible.
Side note: you may have heard of Jeremy Howard in the context of data science or machine learning. Yes, this is the same Jeremy Howard.
So let’s look at what the Ogata study shows and examine the claims made by Manor and Howard.
The Ogata study, which measured free spike protein in plasma
For this study, thirteen healthcare workers with no known history of SARS-CoV-2 infection were given 2 doses of the Moderna vaccine, 28 days apart.
Plasma, which is the liquid portion of the blood, was collected at several timepoints following the first and second jabs.
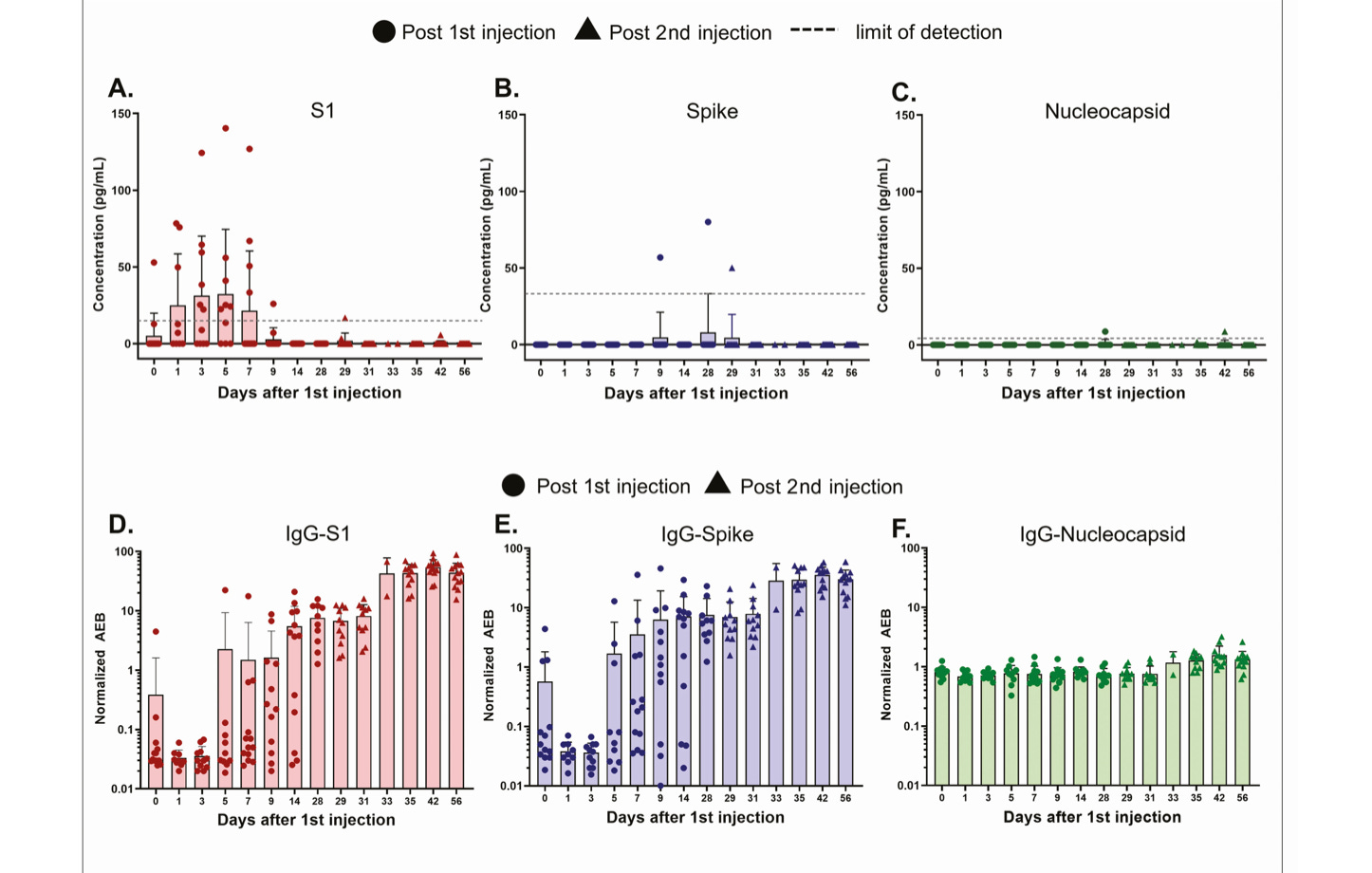
For each time point they measured the levels of the spike protein, and S1, which is a subunit of spike.
In the study, they also measured levels of nucleocapsid, a protein for viral genome packaging in SARS-CoV-2. This was done to rule out the possibility that the S1 or spike that they were measuring in these patients was from viral infection.
People who were infected with SARS-CoV-2 should have been exposed to S1 or spike, as well as nucleocapsid, whereas people who have gotten the vaccine but never been infected, should only have been exposed to S1 or spike.
The red circles show S1 subunit of spike and the blue circles show spike.
Nucleocapsid levels (green) in these patients were undetectable or at “background levels,” which is what you would expect for people who have not had SARS-CoV-2 infection.
It’s unclear what the origin of the spike or S1 subunit in the plasma is. Recall that when the vaccine is injected properly (intramuscularly), it is supposed to get entirely taken up by muscle cells, which are then said to be “transfected.” The mRNA in the vaccines encodes for the spike protein, so the muscle cells should produce spike protein using the newly introduced mRNA as “instructions.” This spike protein is supposed to then get presented onto the surface of muscle cells, where they are supposed to stay attached.
It’s possible that some of the spike or S1 subunit in the plasma are a result of transfected cells that were lysed open upon death. Also free proteases floating about, which are enzymes that cleave proteins, can cleave the spike protein, which has a cleavable S1-S2 site; this could be a source of free S1 subunit floating around in plasma.
In the Manor and Howard article, they say that this study shows that these spike protein levels are “approximately 100,000x lower than physiologically relevant concentrations, let alone pathological.”
Moreover, they say that these levels are extremely small in comparison to the levels of spike protein one would get exposed to under viral infection.
I’ll respond to the first point about sensitivity and levels of spike protein in vaccine recipients in this post (Part 1), and respond to their comparison with viral infection in the next post (Part 2).
Part 1- On Sensitivity and spike protein levels in vaccine recipients
When Manor and Howard say that the Ogata study uses an “ultra-sensitive” assay to measure S1 or spike protein levels, this is a bit misleading, because it makes it sound like the assay picks up on all physiologically relevant levels of S1 or spike protein.
This is just not true.
In the Ogata study, they used an assay called SiMoA (single molecule arrays) to quantify the spike protein or S1 in plasma. This assay was described in a previous study, also by Ogata, as well as here and here.
This assay is a modification of standard “sandwich” ELISA (enzyme-linked immunosorbent) assays where the protein of interest, aka the antigen, or spike protein in this case, needs to bind to two different antibodies in order to be detected by the assay. The antibodies used, which can be bought on sites like here, are known to bind to the antigen of interest.
It’s called a “sandwich” ELISA because the antigen is sandwiched between two different antibodies:
It’s important to understand what was done to quantify spike, in order to assess the claims made by Manor and Howard.
Here were the steps of the assay:
First “capture” antibodies, which in the picture above are the purple things, are immobilized onto microscopic beads. The capture antibody is supposed to contain a site (epitope) that binds to the antigen, aka our protein of interest (spike). Ideally it is specific only to our protein of interest and won’t bind to other things.
The beads sit in tiny wells. Plasma from the blood, which will include water, salts, and proteins, is added to the wells so that the protein of interest that we want to “capture,” is allowed to bind to the “capture” antibodies.
The beads then get “washed” to get rid of things that only weakly bind to the capture antibodies. After the washes, the hope is that only things that bind strongly to the capture antibodies will be left behind.
A second antibody, or “detection” antibody, which is known to bind to the antigen, but at a different site or epitope, is allowed to incubate with the beads.
There are a series of more washings, to remove any detection antibody that is not bound to anything.
Any beads that don’t have any antigen attached to it should not have any secondary antibodies attached to it; we’re assuming that secondary antibodies don’t bind to the first antibodies. We can use this fact to try to quantify how much antigen is present, because the secondary antibody has an enzyme attached to it, and this enzyme, at least in the case of this assay, catalyzes a reaction that leads to the production of a fluorescent compound.
In the last step we measure the amount of fluorescence coming off of all the wells. The amount of fluorescence will correspond to how much antigen was captured. More fluorescence means there were more secondary antibodies present, which means there was more antigen present.
Now, normally it’s not possible to detect ultra-low concentrations of antigens with a method like this because the fluorescent compounds generated are diffused within a large assay volume, and it’s hard to measure this fluorescence amidst background levels of fluorescence.
However, what makes the SiMoA assay different from your run-of-the-mill ELISA assay, is that it uses femtoliter-sized wells (very tiny wells). This means that the beads are contained within extremely small volumes, such that single enzymes can be detected.
So when Manor and Howard say that the Ogata study uses an “ultra-sensitive” assay, this is technically true in comparison to your standard run-of-the-mill ELISA assays. But that does not imply that the assay as a whole picks up on all the spike proteins in the body.
Here are the spike proteins that would not get picked up by this assay:
Spike bound to antibodies
First of all, any spike that is bound to antibodies in the body, would not get picked up by this assay. When I say “antibodies in the body” I mean antibodies that the patient’s body produced as part of an immune response to the spike protein- not to be mixed up with the antibodies used for “capture” or “detection” in the assay.
Ogata et al. also explicitly state this: “Our Simoa antigen assays cannot detect antigen-antibody immune complexes.”
This makes sense given what we know about the assay: any antigen needs to bind to both the detection and capture antibodies in order for it to be detected in the assay. Antigen that is already bound to something, might be able to bind to the capture antibody of the assay, but the antibody from the body would get in the way of binding to the detection antibody, and without that detection antibody with its attached enzyme present, that antigen would not get “counted” or detected by the assay.
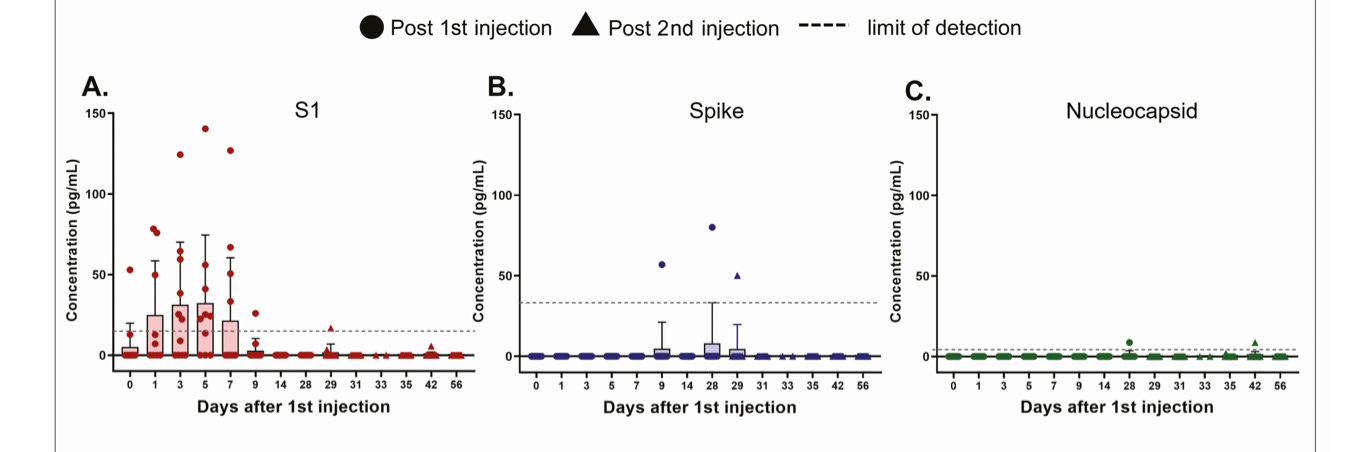
Now, this study also showed that levels of patient-produced antibodies rose initially after injection with the vaccine, and as the levels of these antibodies rose, levels of spike or S1 went down.
Here’s the same image from the Ogata study, but this time also showing levels of various antibodies produced by the patients, where IgG-S1 is an antibody that binds to S1 and IgG-Spike is supposed to bind to spike:
This is often cited as “proof” that the vaccines are “working,” because once the body produces antibodies to the spike protein, it is often claimed that the body has “cleared” the spike.
What does it mean to “clear” spike? Most people would mean that spike proteins that have antibodies attached to them will be “marked” so that components of our immune system can recognize them as needing to be “taken care of,” and those are said to be “cleared.”
However, what we now know is that sometimes this spike protein does not get completely cleared away or “taken care of” by our immune system.
That brings us to:
Spike inside of monocytes, or other cells
In my previous article about the spike protein, in Part II, there is a section called “1. Vax recipients with long-hauler like symptoms.” In this section I discuss the findings of a group of researchers, led by Bruce Patterson. This group found that some vaccine recipients, who had never been infected with SARS-CoV-2, had long hauler-like symptoms for months after receiving the vaccine. In that section I link to a video interview with Dr. Ram Yogendra, who is a part of that research.
These vax recipients were found to have bits of spike protein (S1 subunit) in their non-classical monocytes, which are immune cells that you can think of as “garbage collector” cells. These monocytes, for some reason, were unable to clear the bits of spike protein inside of them, which caused them to release chemicals that caused inflammation. This led to the long-hauler like symptoms, including neurological symptoms.
So sometimes, cells of the immune system, in this case non-classical monocytes, don’t successfully “take care of” the spike protein, even after many months post-vaccination.
The SiMoA assay in the Ogata study would not pick up on any spike or S1 within monocytes, or other cells. Remember, in their protocol they only looked at plasma; this includes water, ions, and free floating proteins, but excludes whole cells. They did not look for proteins inside of cells- in order to do that they would have had to lyse open any cells (break them open) and run the assay on all the crud released from them. That was not the protocol they used.
Spike on the surface of cells or exosomes
Moreover, this assay wouldn’t pick up on any spike protein attached to the surface of cells. That would include any spike protein on the surface of muscle cells that the vaccine transfected, as well as any cells that the vaccine accidentally transfected.
Lastly, a recent study by Bansal et al. showed that spike protein from the Pfizer vaccine could be found on exosomes, which are extracellular vesicles, or packets of lipids that are released by cells. They found that exosomes with spike could be found even months after vaccination.
Given what you know about the assay used in the Ogata study, I hope it’s clear that it would also not pick up on any spike protein on exosomes.
So the amount of spike protein that the Ogata study measured, is likely just the tip of the iceberg, when it comes to physiologically relevant spike in the body.
The bigger point
Here’s the bigger point though. No matter what levels of spike protein one gets from the vaccines, if you believe the findings of the research group led by Bruce Patterson, where they found post-vax patients experiencing negative side effects because of S1 in their non-classical monocytes, we know that the levels of S1 are most definitely physiologically relevant.
Of course you may accept all of this but still claim that it’s worth it to get vaccinated, because it’s better than what you would be exposed to under infection with SARS-CoV-2.
That gets into the other point that Manor and Howard make in their article, where they claim that the amount of spike protein one gets with vaccination is much lower than the amount one would get under viral infection.
That will be taken up in my next post (Part 2).
PS- None of this is to suggest that the level of spike protein in the body is the only relevant factor when evaluating the safety of the vaccines compared to viral infection. There are a myriad of other factors involved, like the delivery mechanism of spike (like virus vs LNPs), as well as differences in the spike protein encoded by the vaccines compared to the spike of the virus, etc. I hope to explore these issues in future posts. You can subscribe or follow here for updates.
Thanks to Christopher M. Brown for useful discussions of the Ogata paper.