What proteins are we actually getting from the COVID vaccines?
As usual, it's not quite what we were expecting
Here’s the story we’ve been told:
The COVID-19 vaccines work by inserting mRNA or DNA into our cells to get them to produce spike protein, which is a protein on the surface of the SARS-CoV-2 virus. Aside from two proline mutations,1 the vaccine-derived spike is basically the same as the SARS-CoV-2 spike.
That may have been what was intended, but there is evidence to suggest that things have not gone as planned.
In fact, we might be getting unknown proteins generated from the COVID-19 vaccines.
Mutant spike pieces in vaccinated people who never had COVID
In July 2022, a paper came out that should have gotten a lot more attention than it did: SARS-CoV-2 S1 Protein Persistence in SARS-CoV-2 Negative Post-Vaccination Individuals with Long COVID/ PASC-Like Symptoms
This paper looked at people who had not been previously infected with SARS-CoV-2,2 had gotten vaccinated,3 and had ongoing long COVID-like symptoms for more than 4 weeks following vaccination.4
They took 14 of these individuals and screened them for the presence of spike protein in a subset of their white blood cells. They found that at least 13 of the 14 patients had spike in their cells.
Of those patients, they chose six patients to isolate spike protein from their cells and determine its amino acid sequence. Remember: all proteins are made up of a string of amino acids. We know what the amino acid sequence of spike protein from the vaccines should be, because we know what the COVID vaccines are supposed to encode for.
What they found was unexpected.
All six of the patients had mutant spike sequences, specifically in the S1 subunit of the spike (the other subunit is S2). In other words, the S1 in these patients had amino acid sequences that were not intended by the vaccine manufacturers.

Perhaps this was due to errors that occurred while mRNA was translated into amino acids. After all, the mRNA of the COVID vaccines had all their uridines replaced with N1-Methylpseudouridine, and there’s literature that suggests this can lead to errors in translation (see here and here).
But the cells of these patients had S2 subunits as well, and those did not contain mutant sequences. Why would we have translation errors in S1 but not S2? Just a coincidence? Or something to do with the order in which the mRNA transcripts are read?
Another possibility is that the vaccine mRNA had errors, and something about the vaccine mRNA manufacturing process caused more errors in the part that encodes for S1 compared to S2.
At any rate, this isn’t good.
All of these patients had long COVID-like symptoms, which presumably were from the vaccines. Would we also find mutant peptides in some vaccinated people who didn’t have long COVID-like symptoms?
It would be great if some researchers would follow this up and sequence spike from more vaccinated people to answer this and get a sense for how prevalent it is. The window of opportunity for doing this is closing, since we'd ideally do this with people who never had COVID.5
The sizes of the proteins we get from the vaccines don’t quite match with what’s predicted
We know what the predicted size of the spike protein should be. So we should expect that vaccine-derived spike would be the same size as what’s predicted for the spike protein, right?
Luckily, there is a very standard technique in molecular biology that allows us to check that: the western blot.
Here’s how it works. You get cells to take up the vaccine and get them to express spike. Then you lyse (break) them open to get their proteins out. You then use gel electrophoresis, which separates proteins by size; proteins with higher molecular weight end up closer to the top of the gel and smaller ones closer to the bottom.
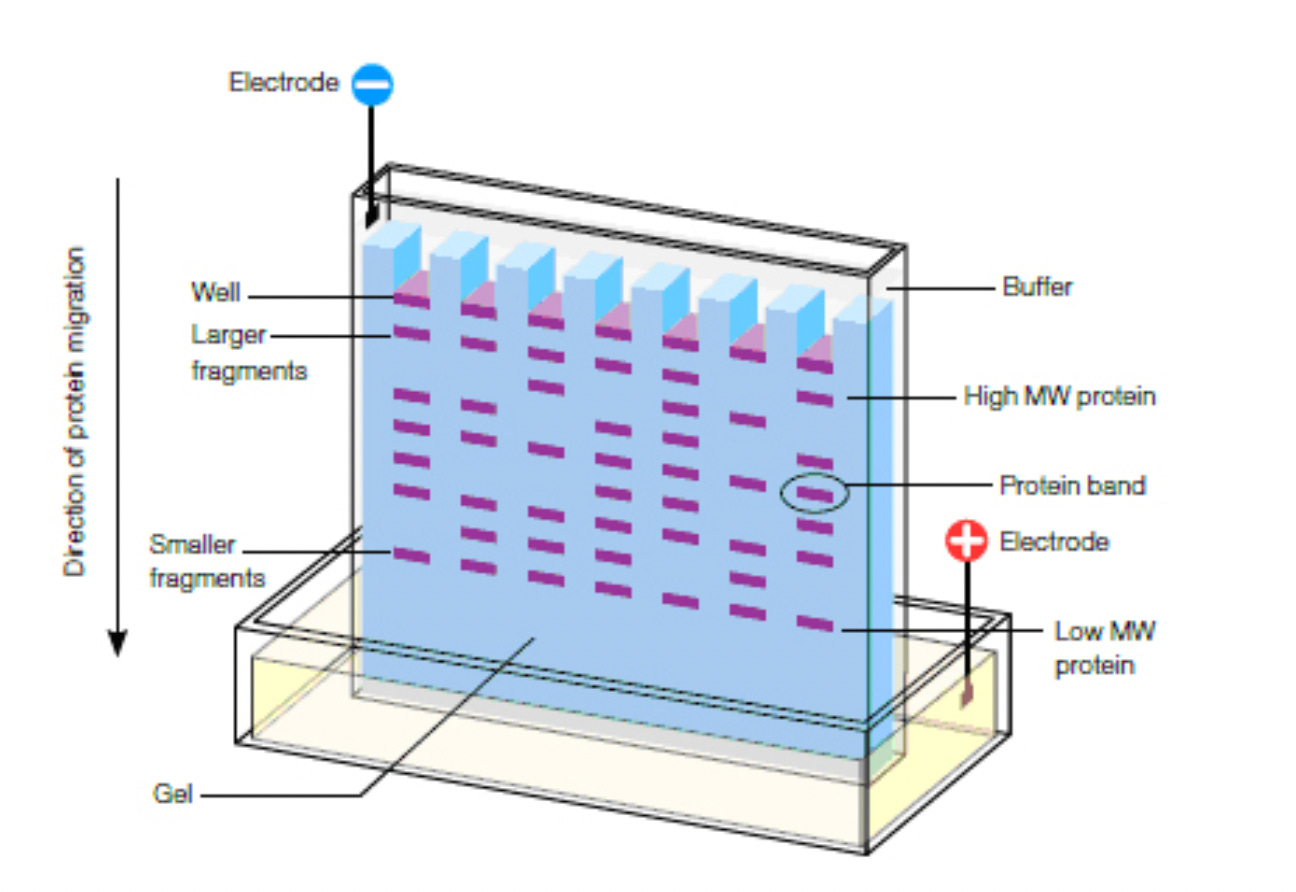
Then the proteins are transferred onto a membrane in a way that preserves where they were on the gel, with larger proteins near the top and smaller ones near the bottom.
The membrane is then incubated with enough antibody specific to spike (“anti-spike”) so that all the spike gets bound to an antibody molecule. We then incubate with a secondary antibody that binds to the first (anti-spike) antibody. This secondary antibody has some property that allows us to visualize it.
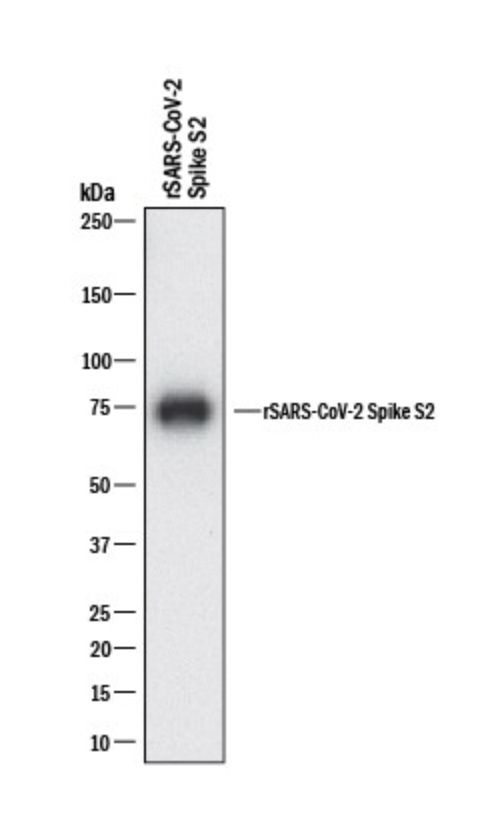
In the end, the spike protein will look like a band on the membrane, and molecular markers can tell us roughly what size the protein is based on where the band is on the membrane. Here’s an example:
References to western blots by vaccine manufacturers
Unfortunately, I couldn’t find any images of western blots performed by the vaccine manufacturers, though we know they were performed.
According to an “Assessment report” for the Comirnaty (Pfizer-BioNTech) vaccine there are references to western blots (emphasis mine):
The expressed protein size after in-vitro expression of BNT162b2 active substance was determined and the results demonstrate comparability between batches. However, the identity of the bands identified by WB are not sufficiently justified and further clarification is requested.
“WB” here stands for western blot.
Further in the document they say (emphasis mine):
Additional data for the active substance are to be provided to confirm the identities of the observed Western Blot (WB) bands obtained by the in vitro expression assay. Protein heterogeneity, resulting in broad bands on the WB and uncertainties in the theoretical intact molecular weight of the spike protein, is assumed to be due to glycosylation... Correlation with the calculated molecular weights of the intact S1S2 protein should be demonstrated.6
Translation: if we're looking at one protein of a particular weight, the band should look relatively skinny and well defined.7 A “broad band” indicates that you might be getting a variety of proteins of slightly different weights, aka "heterogeneity." In this case, they speculate that they got broad bands because of “glycosylation” of the spike protein, meaning it was covered in glycans (carbohydrate molecules), which would add to its molecular weight.
We'll talk more about glycosylation later, but it's too bad we don’t know more about what those bands were.
Luckily, there are a handful8 of studies that show western blot data with vaccine-derived spike. However, they’re not that reassuring.
Western blot with spike from the Moderna vaccine
One of them is a preprint that came out in March 2022: In vitro Characterization of SARS-CoV-2 Protein Translated from the Moderna mRNA-1273 Vaccine
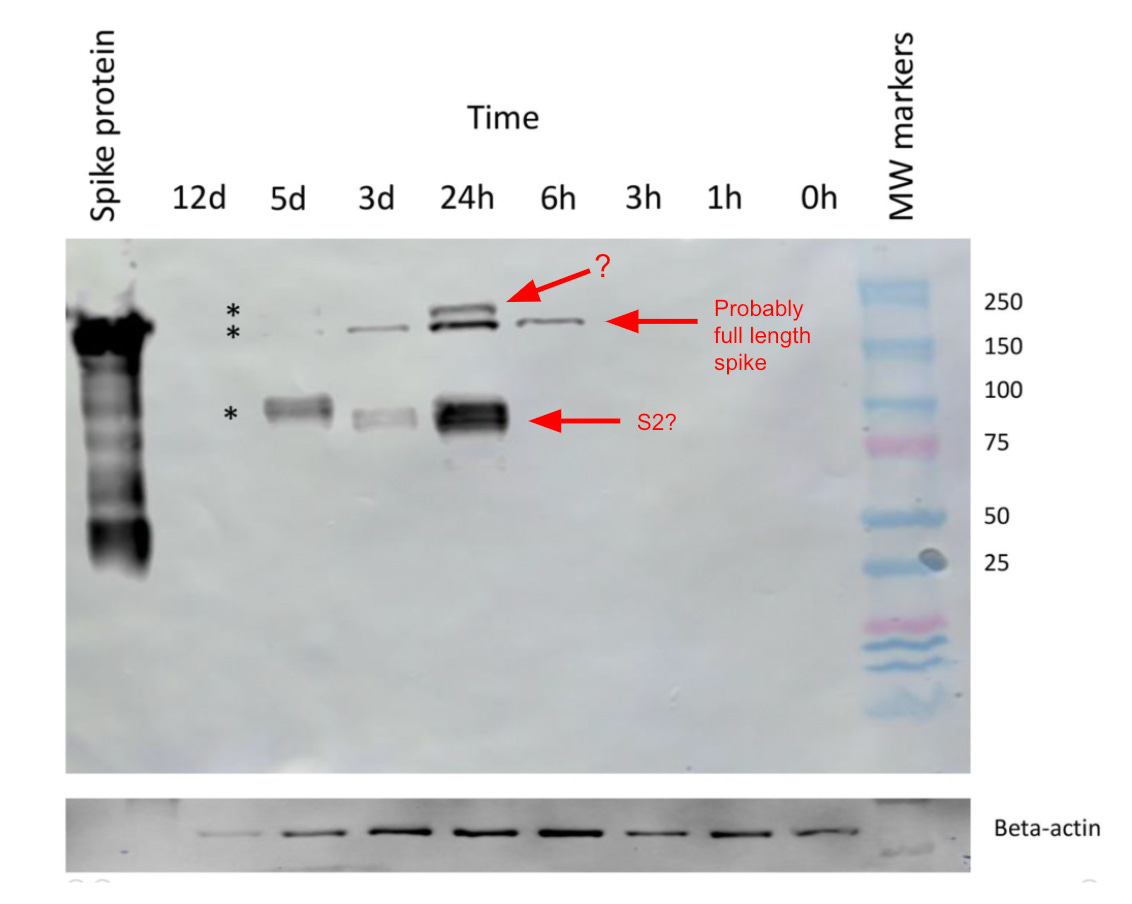
For this study, cells were made to take up the Moderna vax and express spike protein. Then a western blot was performed with protein from those cells.
These are the results9 (the red text is my commentary):
The different timepoints are the times post “vaccination” of the cells, aka, the time since the cells had been given the vaccine.
In this study they used an antibody that binds to the S2 subunit of spike, so I could understand seeing two bands; one for full length spike, and another for just S2, since full length spike can get cleaved into S1 and S2 in the cell by endogenous enzymes.
The lane on the right shows molecular weight (MW) markers. Using that, it looks like we’ve got a band at ~180 kDa that appears at 6 h, 24 h, and 3d; this could be full length spike. That’s because according to this, on a gel10 the full length spike should be ~140-180 kDa.
Then there’s a smaller molecular weight band seen at 24 h, 3d, 5d that looks to be ~100 kDa. I’m going to guess that’s S2.11
But there is a third, higher molecular weight band that appears only at the 24 h mark. What’s that?
Again, is it just glycosylated spike? Remember, glycosylation adds carbohydrate groups to a protein and would add to its molecular weight. But then why would we have two distinct bands? Even if we got spike proteins that were in different stages of glycosylation, with some more glycosylated than others, wouldn’t we expect to see more of a smear, rather than two distinct bands?
Or maybe that band is dimeric or trimeric spike,12 although it doesn’t look large enough. Or maybe it's trimeric S2. Maybe.
By the way, here’s another blot they include, which shows even more bands:
Now, the blots from this paper could definitely use some improvement,13 and it's possible some of the extra bands in this second blot are artifacts.14 But at the least, the higher molecular weight mystery band that’s especially prominent at 24 h seems real, and needs explaining.
It’s unfortunate that we don’t have more to go on than these blots. But as far as I know, this is the only publicly available western blot with Moderna-derived spike.
The authors of the paper mention this (emphasis mine):
In communications with Moderna and Pfizer-BioNTech regarding the proteins expressed by their synthetic mRNA vaccines, each company’s medical information group disclosed that that they had not examined the protein dynamics more than 48 hours post-transfection in cell culture. Owing to its proprietary status, they would not disclose any information related to the nature of the protein that was expressed.
Hmm. What’s “proprietary” about the protein expressed? Weren’t we told exactly what it should be?
Western blot with spike from the Pfizer vaccine
In Nov 2021, a study by Bansal et al., found that people who had been vaccinated with the Pfizer-BioNTech vaccine had exosomes expressing spike protein. Exosomes are membrane-bound sacs that are found outside of cells, like in blood plasma.

In this study they ran western blots of the exosome proteins.
This was from exosomes taken 14 days after the second dose of the vaccine, with each lane coming from a different vaccinated individual:
Unfortunately they cropped out the part of the blot that has the molecular weight markers, but I’m going to guess that this is the full length spike15 that they're showing, using an antibody that picks up S2.
The main point though, is that these look like they could be multiple bands, or broad bands, indicating “protein heterogeneity” as was mentioned earlier in the Comirnaty assessment report. In other words, it looks like a bunch of proteins of different weights.
I hope someone repeats these experiments, and sequences the proteins in the bands (or smears) they see.
Cryptic splicing in the adenoviral vector-based vaccines
The adenoviral vector-based vaccines from Janssen (Ad26.COV2.S) and AstraZeneca (ChAdOx1-S) work by delivering DNA that encodes for spike.
Briefly, here's how they're supposed to work: once the vaccine is inside cells, the DNA is released and enters the nucleus16 where it is transcribed into mRNA. The mRNA then needs to get exported out of the nucleus where it can get translated into protein, but before it does, it can undergo RNA processing, including splicing. Splicing is a naturally occurring process by which the RNA can get cut up in different ways and recombined.
This study (1) predicted sites of splicing in the sequence for spike protein, and (2) found that cells made to express spike also expressed unexpected pieces of protein17 or mRNA18 that appeared to be the result of splicing.
The study also showed that the abundance of splicing events was cell-type dependent, meaning different tissues might produce different proteins from the vaccines.
Moreover, recall that these vaccines are supposed to be injected intramuscularly, and once muscle cells express spike protein, the spike is supposed to remain anchored to the surface of those muscle cells.
According to this study, one product of unwanted splicing is spike protein that lacks the transmembrane domain anchor, which is the part of spike that’s supposed to keep it anchored to the surface of cells. This leads to secreted soluble spike that can float around all over the body. Oops.
If you’re wondering whether this process could occur with the mRNA vaccines, see this footnote:19
Differences in glycosylation are predicted
Glycosylation was mentioned earlier, but it’s worth discussing more.
Spike protein is glycosylated, meaning it has glycan (carbohydrate) groups attached to it. The glycans help the virus hide from the host’s immune system.
In the image below, we see side and top views of spike. The mossy-looking things are glycans. The parts of the protein that are exposed are colored according to antibody accessibility, with black being the least and red being the most accessible.

I don’t know of any studies that checked whether the glycosylation of vaccine-derived spike was the same as in the viral spike. But our default assumption should be that they would be different (see here or here).
Moreover, studies have shown that glycosylation of a protein differs depending on what kind of cell it’s expressed in (see here or here). If that’s the case, why would we expect that vaccine-derived spike, which is supposed to be expressed in muscle cells, would get glycosylated the same way as spike generated from virally infected cells in the nose or lungs?
Maybe it doesn’t matter much for immunogenicity or safety, but then again, the glycans affect which parts of the protein get recognized by the immune system, as well as how the protein is folded or shaped (see here and here).
Differences in protein conformation are predicted
The COVID-19 vaccines used extensive codon optimization in order to increase protein translation. Although this shouldn’t change the amino acid sequence of the protein expressed (theoretically), it can affect how a protein folds, or its 3-D conformation (see here or here). This in turn could alter how it functions.
This review explores how the modifications to the mRNA might have led to unexpected consequences. One obvious alteration is that the codon optimized mRNA vaccines have considerably more guanine and cytosine compared to the RNA from the virus.

This could alter mRNA secondary structure and lead to something called G-quadruplex structures. It’s an open question whether this affects translational efficiency or creates more errors or truncated spikes.
This technology is not even close to mature
We were told that the vaccines yield spike protein, but we’re not even sure whether very basic things, like the sizes of the proteins from the vaccines, match with what’s expected.
Here’s a list of all the different proteins we might be getting from just the nucleic acid portion of the vaccines (so not even including things like LNPs, etc):
The predicted spike protein
Pieces (subunits) of spike that are the result of cleaving spike by host enzymes
Proteins that are the result of errors in translation
Proteins that are the result of errors in vaccine manufacturing or handling. Some analyses, by the way, have correlated more adverse reactions with certain vaccine lot numbers.
Proteins that are the result of splicing or hidden genes. More on hidden genes here: Are There Hidden Genes in DNA/RNA Vaccines?
Proteins that have the same sequence of spike but have different folding or 3-D conformations
Variations of all of the proteins above that have different glycosylation
That’s a lot of different possible proteins.
Some of these errors might even partially explain why some people experience serious adverse reactions from the vaccines while others don’t. We don’t know.
Whatever is happening, the fact that we might not even be getting consistent product from the vaccines makes it difficult to get at the specific mechanisms of action behind adverse reactions.
You may have heard that research into nucleic acid vaccines has “a long history.” That is misleading, at best. We can’t say that this technology is even close to mature, when we don’t even know what we’re getting from them.
I’d like to thank Christopher M. Brown for useful discussions interpreting the western blot with Moderna-derived spike.
This would stabilize it into its prefusion conformation. This was done in the Pfizer, Moderna, and J&J vaccines to make the spike more immunogenic. More here.
Lack of prior infection was determined by looking for anti-nucelocapsid antibodies or a T-Detect Test. I don’t know how accurate these tests are, but it’s of course possible that at least some of these patients had unknowingly been infected at some point and the mutant spike protein was from viral infection, not the vax. Either way, the data from this study needs explaining.
Participants in the study had gotten either the Pfizer-BioNTech, Moderna, Johnson or Johnson, or Oxford/AstraZeneca vaccine.
On average, symptoms were reported 105 days post-vaccination, though at least one individual had symptoms 245 days post-vaccination (Table 1).
It would need to be done in people who haven’t had COVID, otherwise we wouldn’t know whether any spike found was from the vaccine or virus.
The vaccine manufacturer was asked to demonstrate that they get the expected bands with a due date of July 2021. Unfortunately I haven’t been able to find the documents that contain this.
[UPDATE 9/30/22: According to these newer EMA docs the obligation to show the correct bands was completed. On p.19 it says “WB restults obtained by three different antibodies, specific for the S1 domain, the receptor binding domain and the S2 domain, respectively, were presented and compared to theoretical masses of the S-protein and the subdomains in glycosylated and non-glycosylated forms. It is sufficiently justified that the major band monitored corresponds to the heavily glycosylated S-protein.”
My thoughts: this doesn’t actually show the blots, so we don’t know how they were able to determine that what they were looking at was glycosylated spike. It also ignores the fact that if they are only using antibodies for S, S1, or S2, they won’t find proteins that are sufficiently different from spike.]
Sometimes it might still look broad if there is a lot of protein there, but even in that case it should look well defined. If it looks like a smear, that might indicate that the band might be composed of proteins that are heterogeneous.
There’s one other study that I know of that performed a western blot with vaccine derived spike and it’s this one: Fertig et al. I don’t discuss it in this article because no visible spike could be seen in their blots (Fig. 3). This makes me wonder whether the vaccine just doesn’t “work” as well in certain cell lines.
Something that’s also interesting in this figure is that it seems like beta-actin expression goes down sometime after 3 d. Beta actin is often used as a loading control in western blots to normalize the levels of protein detected because it’s thought to not change much across conditions, but we can see here that it goes down (assuming they are loading equal lysate in each lane). Is overall protein in the cell going down?
This is when using SDS-PAGE under reducing conditions. Note: the predicted size of this protein is a bit less than what you see in gels because of things like added sugars from glycosylation.
Different sources vary on the molecular weight of S2 you’d expect from a gel under reducing conditions. Here a recombinant S2 protein (with a His-tag attached, which adds ~1 kDa) appears to be over 100 kDa, although for some reason they said that the blot was “showing bands at 65-75 kDa” (it doesn’t; the bands are clearly much higher up than that). Here in the first western it looks like S2 is 70 kDA whereas the second western ("simple western") shows it being ~91 kDa. So all I can say is that S2 appears to be somewhere between 70-100 kDa.
Meaning two or three spike units joined.
The reason I say this is that in Fig 2 and the first blot of Fig 3, their spike protein positive control (in the leftmost lane) is just a huge smear (this might have happened because they loaded too much protein, and the spike protein might have been insufficiently purified). A positive control in this context is a lane with pure protein that you’re highly confident is the protein you’re looking for, in this case spike; you include this because if you don’t see anything in this lane you know something went wrong with your blot. Then in Fig. 3 they don’t show any spike protein at all, including from their positive control, in the U937 cells. In that case, something went wrong with the blot, since we don’t even see the positive control. My guess would be that reviewers would ask the researchers to do these over again.
Like if there was too much antibody or too much protein loaded onto the gel or if the sample has degraded, you can see numerous bands like this.
If you’re wondering why we would only see full length spike and not S2 in this blot it might be because the only spike they get from exosomes are full length spike (those are the only ones attached to the surface of exosomes). It doesn’t mean that these patients didn’t also have S2 units somewhere in their body; just that it wasn’t found associated with exosomes.
This shouldn’t be confused with the spike protein itself getting into the nucleus, which I previously wrote about here.
One of the things they did was transfect cells with a “splice trap construct” which encoded for spike protein upstream of a fluorescent protein (luciferase) missing a start codon. The way this was constructed was such that we’d only see fluorescence if splicing occurred. Fig. 2 shows fluorescence activity from cells, indicating splicing activity. These cells also made proteins of many different sizes, according to western blots (Fig. 3). Though this is not the same as showing aberrant bands coming from cells that had taken up the actual vaccines, it suggests that the sequence for spike (which the vaccines encode for) has splicing sites.
One way they showed this was that they took the mRNA from cells that had been transduced with adenoviral vectors encoding for spike and then constructed cDNA (complementary DNA) from the mRNA, then used PCR to amplify segments of the cDNA. This resulted in some DNA fragments that were different from what we’d expect based on the primers used, suggesting splicing had occurred at different positions.
In order for this to be occurring with the mRNA vaccines, I assume the vaccine would first have to get into the nucleus, which is where splicing occurs. My assumption is that it’s possible for the LNPs to get into the nucleus based on some literature (example here) describing the use of LNPs to target the nucleus (though of course not all LNPs are the same). However, even if they got into the nucleus, it’s unclear whether the mRNA would get spliced or modified the same way as normal mRNA would; the vax mRNA had extensive modifications to it, including replacing uridines with N1-Methylpseudouridine. In short, I don’t know whether this is possible with the mRNA vaccines, but I don’t think we can rule it out entirely either.





Wow, I didn't even think of the fact that muscle cells are being used to express the spike and what factor that would play, but given your previous post on the HeLa contamination this should raise a lot of questions. Now I'm wondering if the multinucleated nature of muscle cells may actually be an issue in both gene expression and uptake of even things such as the adenoviral vectors. Lots more things to ponder and it raises even more questions! When it comes to glycosylation do you suppose that the cell lines used for studies would be critical as well, such that in vitro assays may not provide a viable examination of the true glycosylation occurring if we assume that there will be high variability due to different biochemical processes.
But I suppose then that with those with severe COVID and widespread viral infection should we assume differences in protein conformation and glycosylation? Sorry This whole post has made me really curious about everything!
The fact that the actual spike protein from the vaccine hasn't been fully elucidated is rather alarming as well. Do you know how the mRNA is constructed Joomi? I generally assumed some sort of chip technology automated with some base insertion/wash sequence, but I suppose the type of method would also raise questions as to the fragments produced and whether they were properly isolated.
What the hell? How could regulators not have asked if the proteins churned out were actually what was expected? Was no sequencing done at all? Or just dodgy blots?